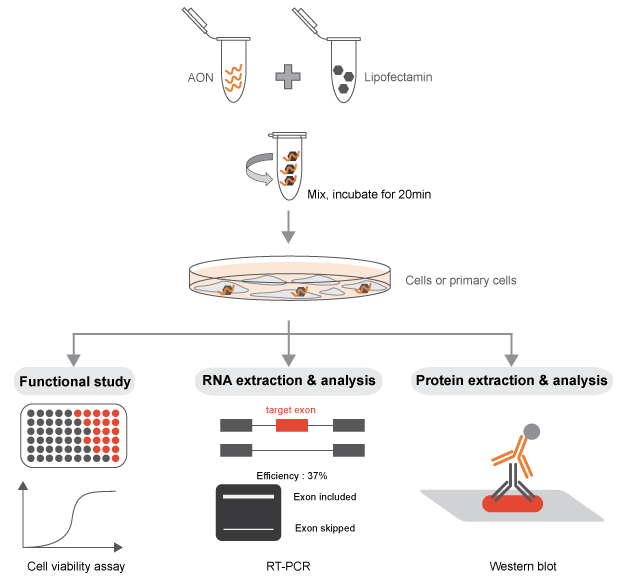
Re-suspending the AONs
The AONs are supplied in lyophilized powder or gel form and can be re-suspended in distilled, sterile water
or PBS buffer. They are very soluble in water.
Storage of the AONs
The AONs must be stored at ≤-20 ºC. When stored in powder form, the AONs are very stable for more than 1
year. Once re-suspended in water or buffer, it is recommended to aliquot them and use within 1 year. It is
recommended to prepare small aliquots in order to avoid repeated freeze-thaw cycles.
Use of the AONs in vitro
In order for the 2-O-Methyl phosphorothioate AONs to be functional in the alteration of a specific splicing
event, they must be able to access the nucleus of the cell, where the splicing reaction takes place. The
AONs synthesized with this chemistry are capable to self-penetrate the cell (and the nucleus) when added to
the media in multiple cell types. However, we recommend using transfection reagents or electroporation that
will allow to achieve the desired splicing switch with a minimal amount of AONs in in vitro experiments. A
titration of the AON concentration is highly recommended for all the experiments performed.
Naked AONs
Naked AONs are simply added to culture medium once re-suspended in water or PBS buffer. It must be taken in
consideration that the naked AON uptake is cell-type specific: cells with “endocytic nature” (e.g. APCs)
will be able to naturally have more productive switching, while secretory cells (e.g. T cells) are going to
be less prone to internalize the AONs. Usually, the amount of AON tolerated by cells in the medium spans in
the range of single-digit µM to nM.
Transfection reagents
We noticed that the transfection reagents and protocols used for siRNA or plasmid’s transfection are
working for AON as well, leading to an improved uptake of AON in cultured cells. We noticed a sensibly
better performance for AON’s transfection in adherent cells, such as lung carcinoma A549, and prostate
cancer cell lines PC-3 and LnCap, using RNAiMAX and Lipofectamine 3000 compared to Lipofectamine 2000 (see
Protocols section). With Lipofectamine 3000 as transfection reagent, the IC50 of an
effective AONs can range from 5 nM to 250 nM depending on the cell lines and target genes.
Electroporation of AONs
For cells cultured in suspension, electroporation systems have been shown to suit the AON intra-nuclear
delivery maintaining the same parameters used for siRNA and plasmids.
Use of the AONs in vivo
Our previous study (Lin J et
al., 2017) with mouse xenograph models showed that intraperitoneal injection of the naked AONs
has successfully induced target exon skipping and inhibited tumor growth.
Detection of the splicing switch
Splicing is a constant and dynamic process. The switches in the desired splicing event can be observed as
early as within a few hours from the transfection of the AON. The factors influencing the kinetic and
efficacy of the switch are multiple: transcript abundance, transcription turnover, stability, etc. The
switch can last for numerous days, depending on the target transcript, cell type, and replication rate.
Non-NMD-inducing AONs
If the AON is not meant to induce the degradation of the target transcript, the detection of the splicing
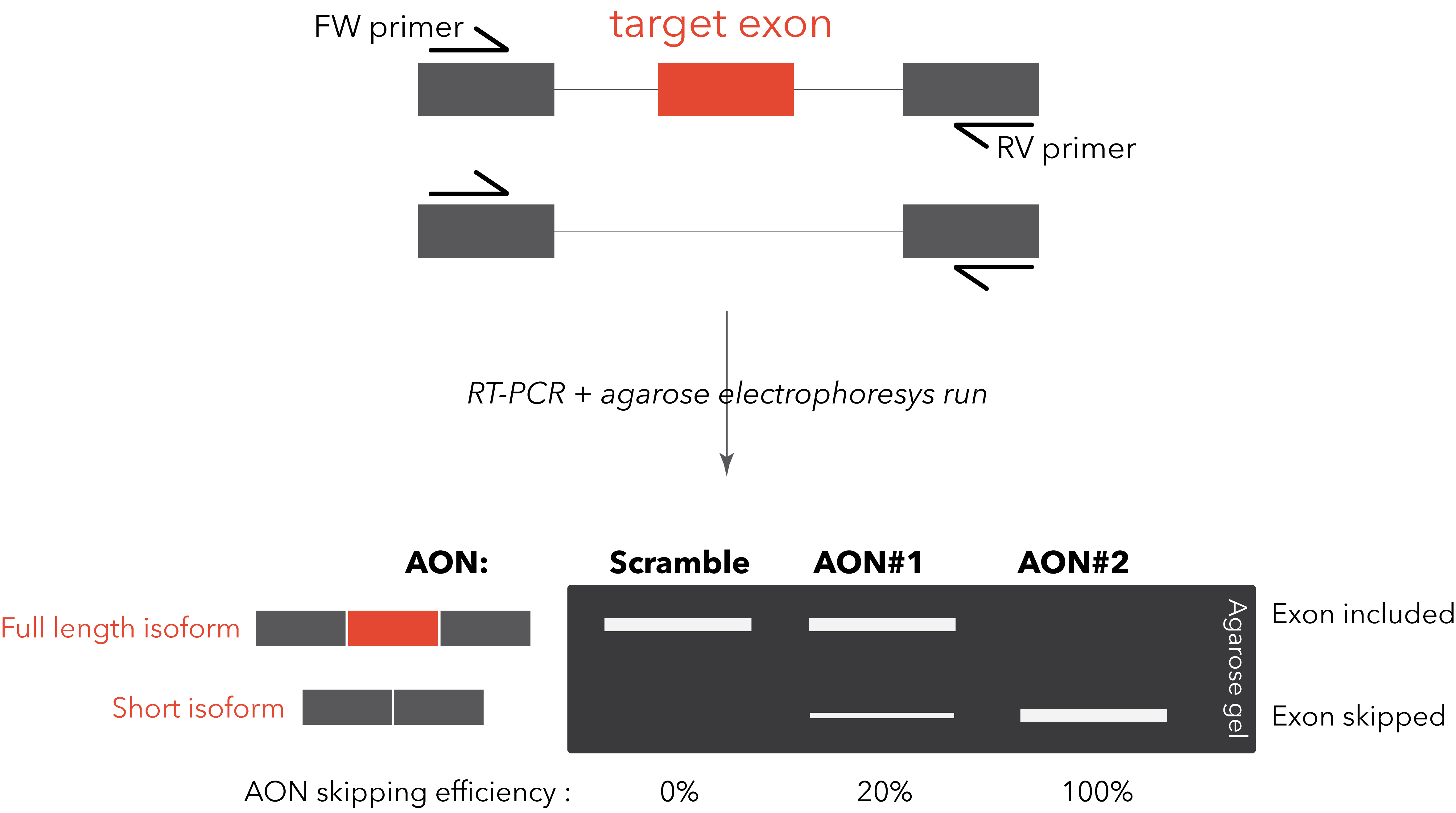
switch is usually straightforward. A simple RT-PCR with primers designed on the flanking exons will be
sufficient to visualize on an agarose gel the two different bands (full length isoform and “skipped”
isoform). This method allows a rapid and clear quantification of the AON efficiency. A TaqMan PCR, specific
for the two isoforms, can be designed for improved quantification sensitivity and reproducibility (Dewaele M et al., 2016).
However, it might happen that the target exon doesn’t have flanking exons that are constitutively included
in the transcript. We suggest to carefully look at the alternative splicing occurring in the surroundings of
the desired splicing modulation. In case the alternative splicing pattern is particularly complex, we
suggest to design the primers on constitutive exons and use capillary electrophoresis to detect and quantify
all the possible isoforms (Tabaglio T et
al.).
NMD-inducing AONs
For AONs designed to induce degradation through nonsense-mediated decay (NMD), the switch will not be
easily quantifiable since the isoform subject to NMD will be degraded. A simple quantitative PCR (qPCR) to
detect the decrease of the target mRNA in this case will suffice to assess the efficacy of the AON switch.
In combination, a treatment of the cells for few hours with cycloheximide before cell harvest will block the
NMD process, allowing a more reliable quantification of the switch.
Troubleshooting if no or low target switching could be detected
Possible reasons and solutions:
- The AONs cannot penetrate the cell (and/or the nucleus).
Solution: If naked AONs was used, try to increase the concentration of AONs or use a transfection
reagent such as lipofectamine. If transfection reagent was used, try to optimize the transfection
conditions. For example, adjusting the ratio of AON/transfection reagent.
- The AONs concentration is too high.
Solution: We have noticed that if the AONs concentration goes too high (e.g. >250 nM) in in
vitro experiment with transfection reagent, their target switching effect may start to go off. Therefore,
we recommend a concentration of 50 to 100 nM for each AON to start with.
- The transcript with the target switching has been degraded via NMD.
Solution: If the transcript with the target switching has been degraded through nonsense-mediated
decay (NMD), no or low switching could be detected. In order to protect the transcript with the target
switching from NMD, cycloheximide (suggested working concentration: 10 – 100 µg/ml) can be added into the
medium 5 hours after transfection.
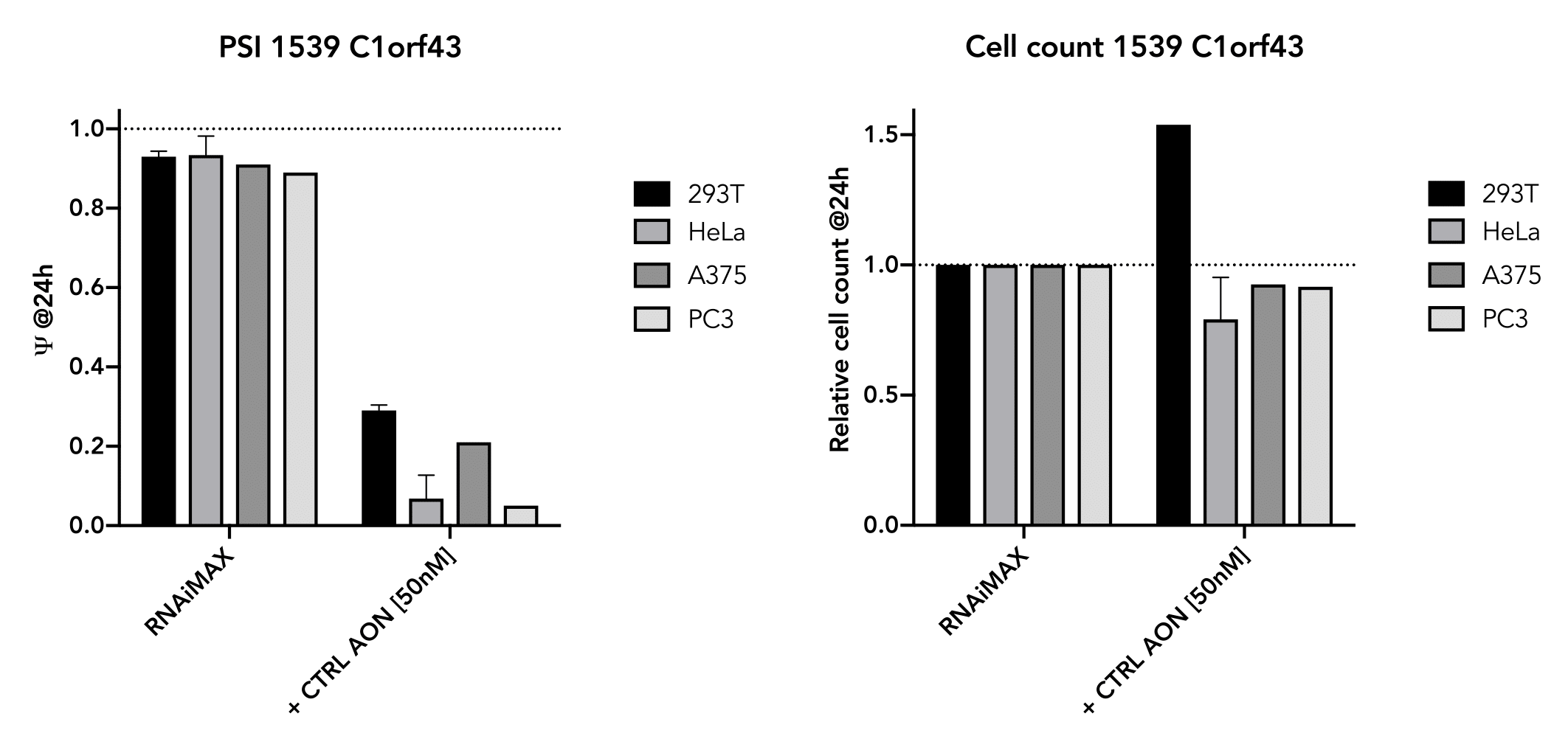
Positive and Negative Controls
The positive control has also been shown to work well in several cell-lines (293T, HeLa, A375, PC3) and
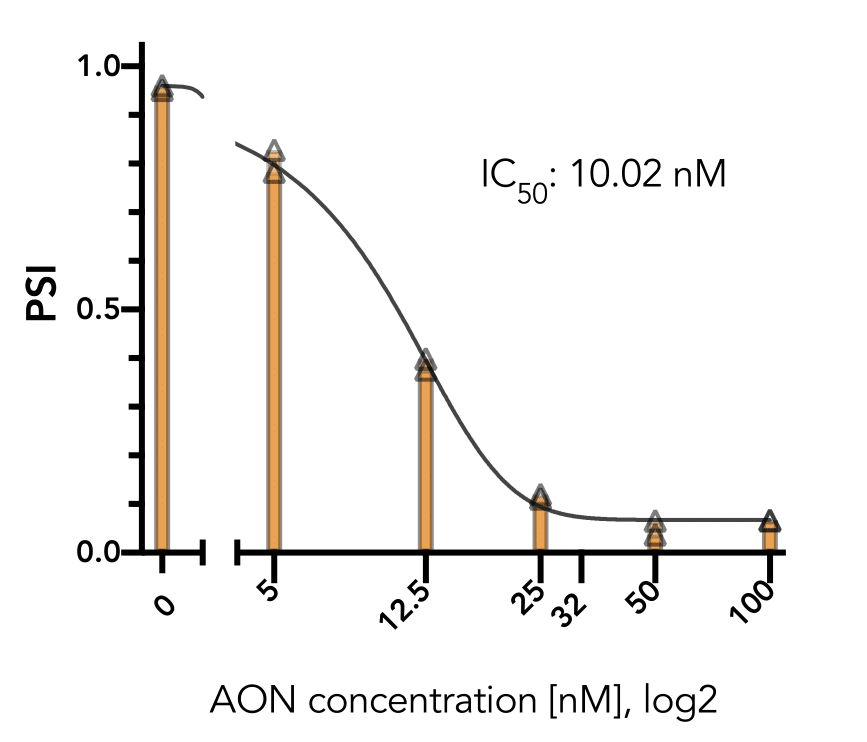
generally do not affect cell viability in the short term (24 hr). The exon-skipping efficiency of our
positive control is high (IC50:10.02 nM in HeLa cells) and works in a nice dose-dependancy manner.
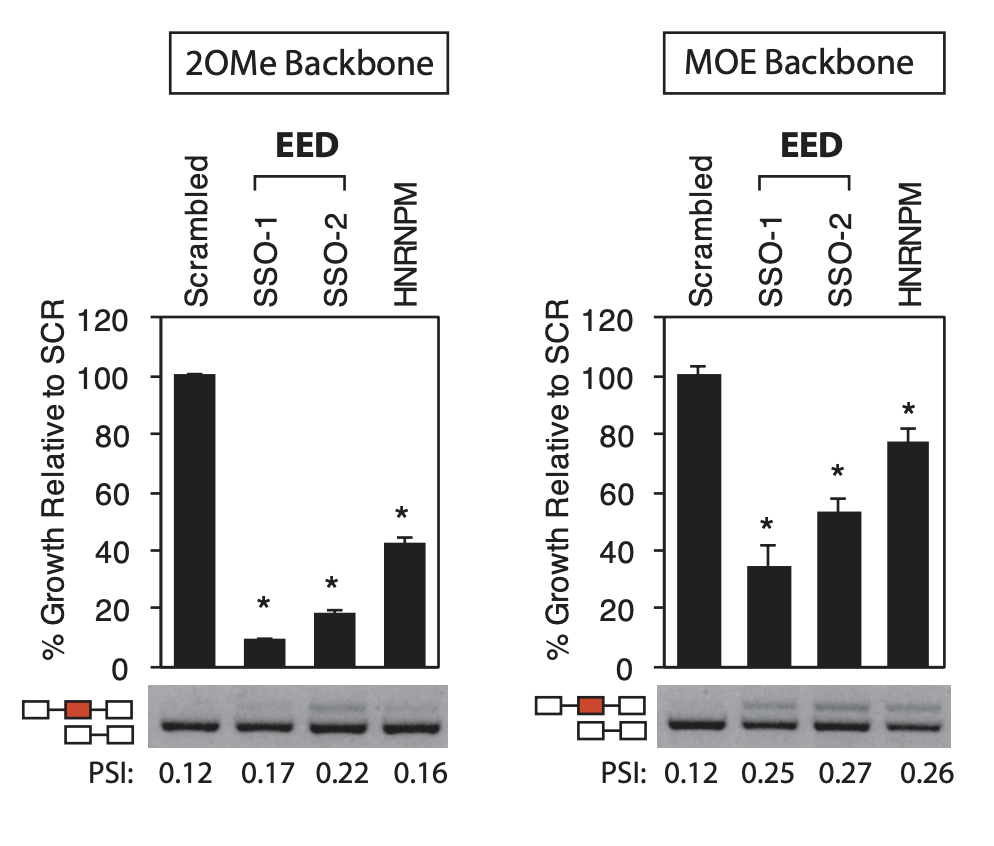
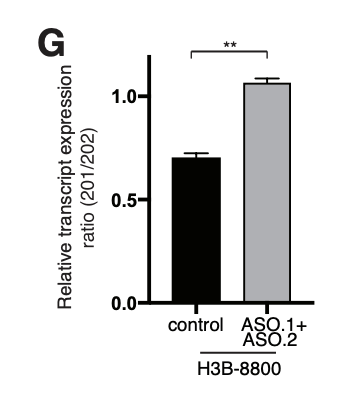
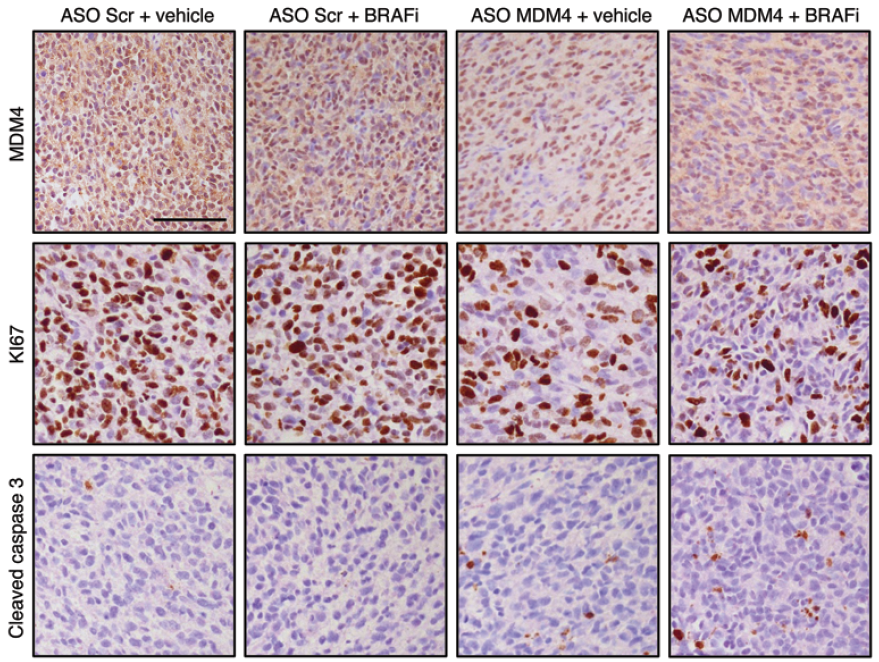
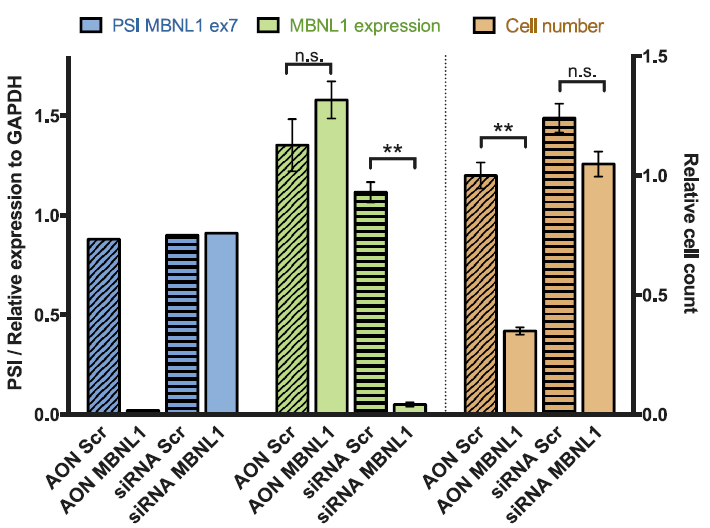
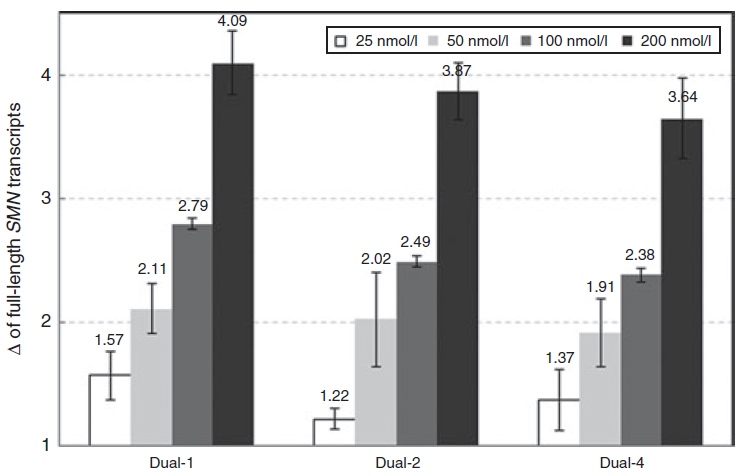
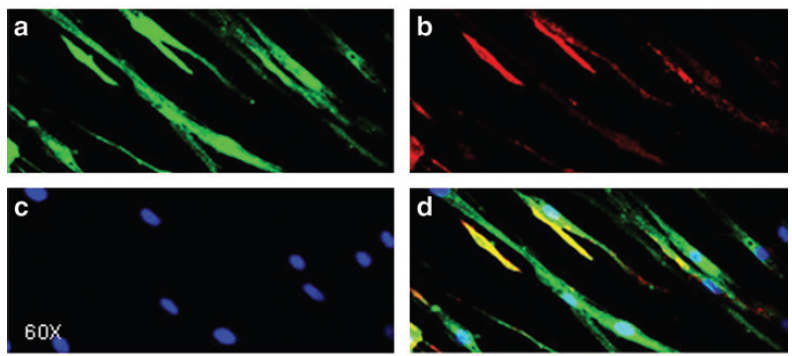
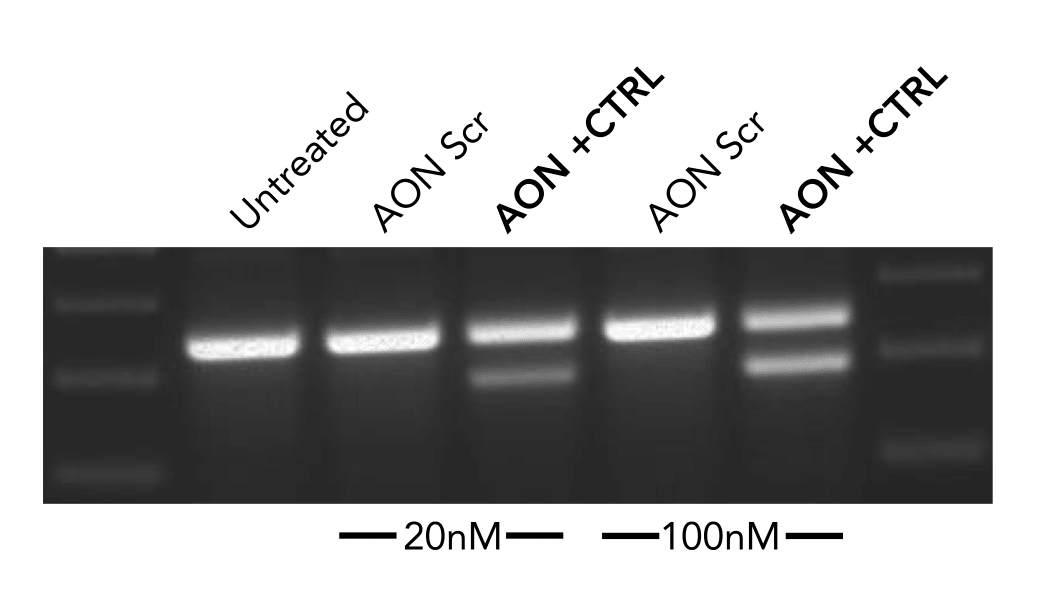
The negative control used in our studies is labeled as "AON Scr" (Scrambled) in the following figure. They
have been tried and tested in many different human cell lines. The figure below shows its efficiency in
skipping an exon in mouse NIH-3T3 cell line.