NOA-GAPMER™
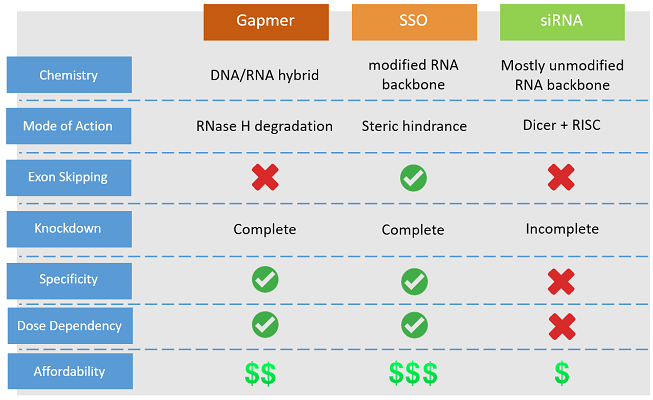
- Function : gene/transcript knockdown
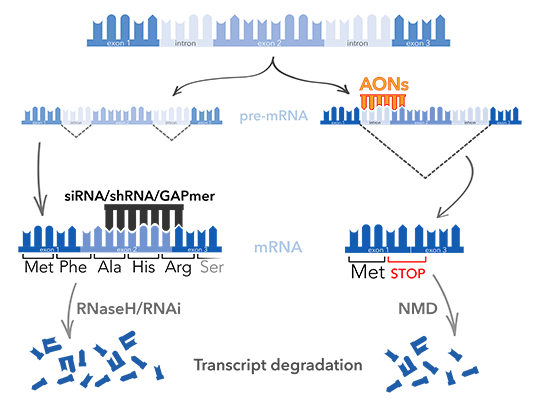
- Action : RNaseH degradation
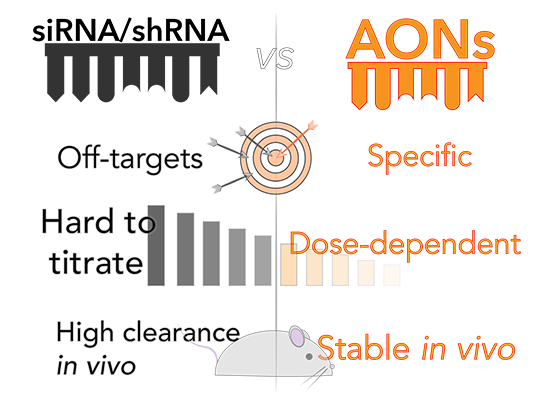
- Advantage : target specificity & dose dependent
- Affordability : significantly more affordable as compared to NOA-NMD™
NOA-GAPmer™ is very efficient in transcript knockdown and exhibit nice dose-dependency.
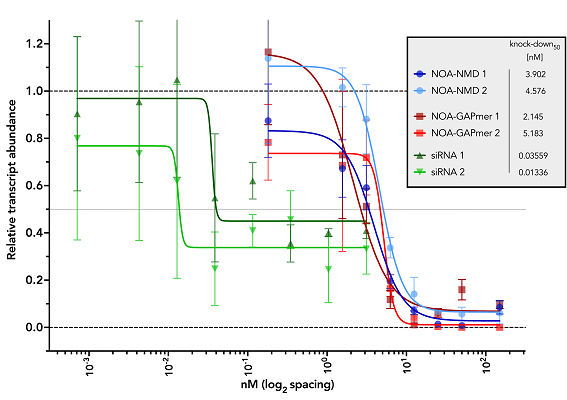
Knockdown levels of GLDC gene upon transfection with two single siRNAs and TechNOA designed NOA-NMD
and NOA-GAPmer AONs. The transfection was performed on A549 cells with lipofectamine RNAiMAX and
RNA was harvested at 24h post-transfection. The qPCR data for GLDC was normalized against the
housekeeper HPRT and the relative non-targeting controls (non-targeting siRNA, non-targeting NOA-NMD
and non-stargeting..
NOA-GAPmer) set at a value of 1. The results shown are average and standard error of
mean of three biological replicates. While NOA-NMD and NOA-GAPmer fully knockdown the GLDC transcript,
increasing concentrations of siRNA was not able to fully knockdown the transcript.